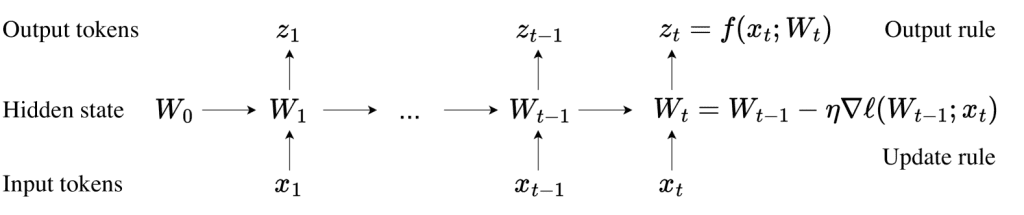
Traditional biological research, often characterized by labor-intensive experiments, struggles to reveal the intricate mechanisms behind protein folding and function. The advent of large neural networks offers a transformative approach by uncovering hidden patterns and making accurate predictions, particularly in protein biology—life’s fundamental code.
Biology is fundamentally programmable. Every living organism shares the same genetic code across the same 20 amino acids—life’s alphabet. — EvolutionaryScale
EvolutionaryScale recently introduced ESM3, an advanced multimodal generative language model designed to simulate over 500 million years of protein evolution. This model stands at the cutting edge of computational biology, integrating sequence, structure, and function data to generate novel proteins, effectively simulating evolutionary processes that would naturally span eons.
ESM3’s Architecture:
The core of ESM3 lies in its ability to handle and integrate three key aspects:
• Sequence Modeling: Using language models to analyze and predict amino acid sequences of proteins.
• Structure Prediction: Integrating sequence data to predict the three-dimensional structures of proteins.
• Function Prediction: Combining sequence and structure data to predict the functional characteristics of proteins.
This architecture enables ESM3 to generate biologically functional proteins, simulate long-term evolutionary processes, and explore previously uncharted areas of the protein space.
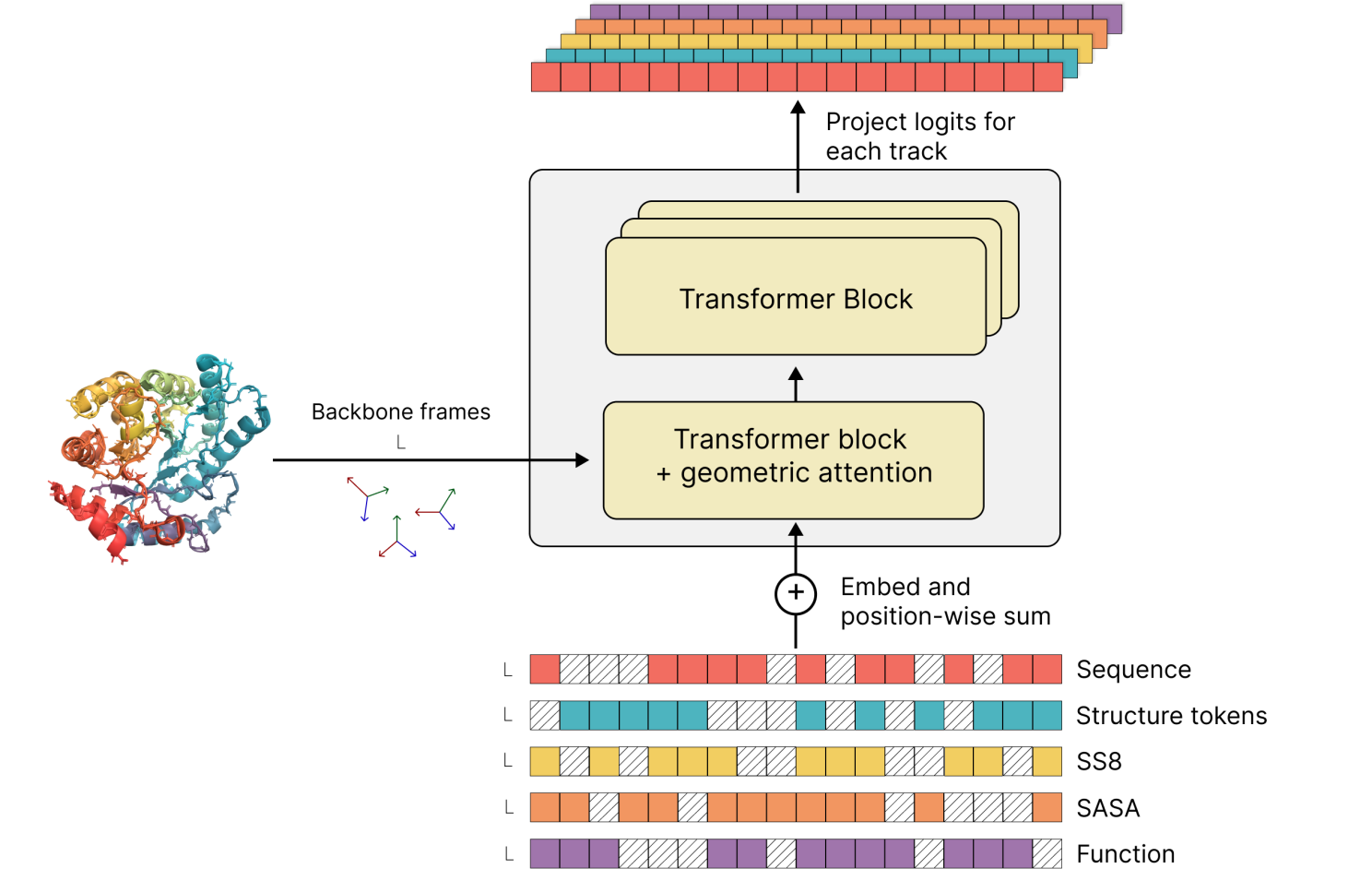

How ESM3 Generates New Proteins:
• Input: The process begins with inputting an amino acid sequence into the model.
• Structure Prediction: The model’s structure prediction module transforms this sequence into its corresponding three-dimensional structure, utilizing vast biological data to predict stable protein folds.
• Function Optimization: ESM3 further refines and optimizes the sequence and structure to generate new proteins with specific, desired functions, simulating mutations and selections that occur during natural evolution.
The research team successfully utilized ESM3 to generate a novel fluorescent protein, esmGFP, which mirrors the characteristics of natural proteins.
Future Applications of ESM3:
Protein Design and Generation
• New Protein Generation: ESM3 enables the creation of entirely new protein sequences, allowing scientists to design proteins with specific functions, a crucial tool in molecular biology research.
Drug Discovery and Development
• Cancer Therapy: By designing proteins that target specific cancer cells, ESM3 could contribute to the development of novel anti-cancer drugs.
• Antibody Design: The model can be used to design more effective antibodies, enhancing their activity against pathogens, which is crucial for vaccine and therapeutic development.
Environment and Sustainability
• Carbon Capture: ESM3 has the potential to design proteins that capture and sequester carbon dioxide, offering a new approach to mitigating climate change.
• Plastic Degradation: By designing enzymes like PETase, ESM3 can play a role in plastic waste management and environmental protection.
Biological Systems Programming
• Synthetic Biology: ESM3 offers a programmable biological platform where scientists can specify protein designs through natural language, driving advances in synthetic biology.
Scientific Research and Education
• Fundamental Research: ESM3 accelerates the study of protein structure and function, helping scientists better understand the fundamental principles of biology.
• Educational Tool: As a powerful educational resource, ESM3 can be used in teaching and training, helping students and researchers learn about protein science and bioinformatics.
ESM3 is not just a technological advancement; it represents a paradigm shift in how we approach biological research and protein design, unlocking possibilities that were once confined to science fiction. The future of biology is now programmable, and ESM3 is leading the way.
Click here to check EvolutionaryScale.